Ppe-Acid: Peach Acidity
Titrable acidity is an important attribute of peach quality. An optimal balance between acidity and sweetness is sought ensuring that peach cultivars are neither bitter due to too much acid nor overly bland due to a low acid concentration. Peaches can be categorized as either high or low acid, although wide variation is found within both. The ability of a breeder to select individuals with ideal fruit acidity is key to the development of new flavorful cultivars.
Genetics
Most genetic variation for acidity observed in U.S. commercial peach germplasm is controlled by two genomic regions, the D locus and the G7Flav locus. A DNA test was developed that targets both of these regions by combining two tests, CPPCT040b* and G7Flav-SSR, into a single, powerful test: Ppe-Acid. CPPCT040b targets the D locus which is responsible for the difference between high (d|d) and low (D|D or D|d) acid peaches. Low acid is dominant to high acid. G7Flav-SSR targets the G7Flav locus that is needed to differentiate acidity levels within the two main acidity groups. One allele, H, increases the acidity within each group and another allele, L, reduces the acidity within each group. Ppe-Acid is a simple PCR-based test consisting of two SSR markers, each targeting a different locus, multiplexed into a single assay.
Predictive Capacity
With just CPPCT040b, the two major acidity groups can be differentiated. By also incorporating G7Flav-SSR, the extremes within the high acid or low acid groups, which are likely too high or too low for consumers’ liking, can be routinely identified. The average titratable acidity (TA) for a low acid peach in commercial U.S. germplasm ranges from ~0.2 to ~0.5 TA (mEq/L) as predicted by CPPCT040b alone. The individuals with very low acidity as predicted by CPPCT040b & G7Flav-SSR have an average of ~0.25 TA while those predicted to be mid-low acidity have an average of ~0.45 TA. The predictive power of these tests combined was confirmed in the RosBREED project on four U.S. peach breeding programs and is now being routinely used. Confirm the effects in your own germplasm before widespread use.
When to Assay
As with all DNA tests, there is benefit to be gained from testing parents for cross selection, but the peach acidity test really shines when used for seedling selection. Many of the allelic combinations of CPPCT040b and G7Flav-SSR are undesirable resulting in acidity that is either too high or too low. With the Peach Acidity DNA testof Ppe-Acid, you can cull inferior seedlings early that would otherwise be planted and grown to maturity over two to three years before being phenotypically tested and eventually discarded!
Allelic State of Several Peach Cultivars
| Genotype | Example Cultivars | Phenotype |
|---|---|---|
| D |- & 278 | 278 | Sugar Lady | Very low acidity |
| D | - & 338 | 338 | Amoore Sweet | Low acidity |
| d | d & 278 | 278 | Contender | High acidity |
| d | d & 338 | 338 | Tropic Prince | Very high acidity |
A table of haplotypes for important U.S. peach germplasm can be downloaded here.
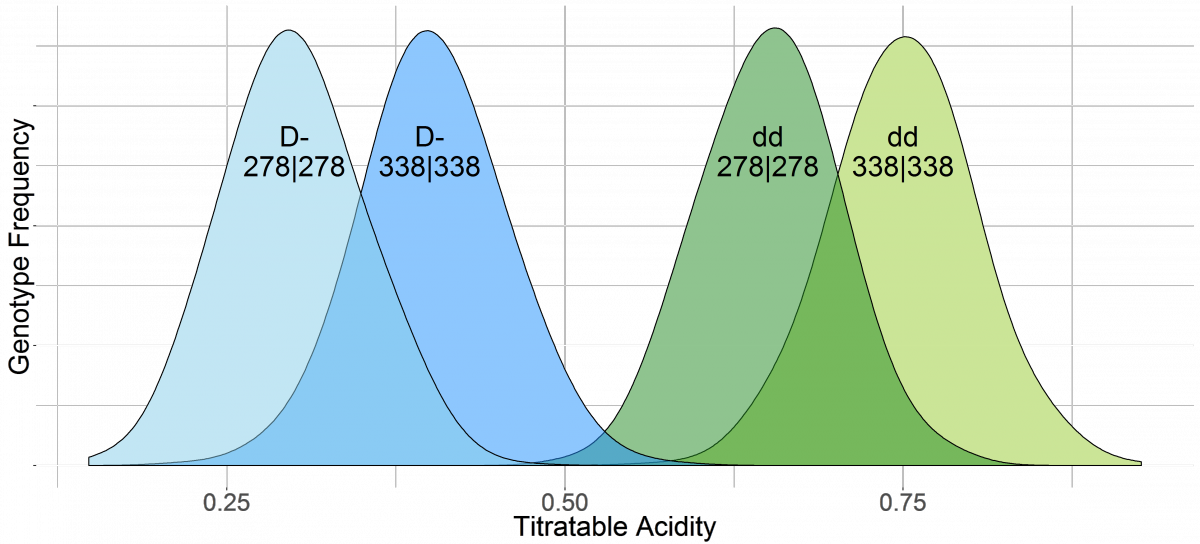
Phenotypic Distribution of Acidity Genotypes
Alleles Available
CPPCT040b targets the D locus which is responsible for the difference between high (d|d) and low (D|D or D|d) acid peaches. Low acid is dominant to high acid. G7Flav SSR targets the G7Flav locus that is needed to differentiate acidity levels within the two main acidity groups. One allele, 278, increases the acidity within each group and another allele, 338, reduces the acidity within each group.
Technical Details
Ppe-Acid is a simple PCR-based test consisting of two SSR markers, each targeting a different locus, multiplexed into a single assay.